colorSV: Detecting Somatic Structural Variation Using Tumor-Normal Co-Assembly Graphs
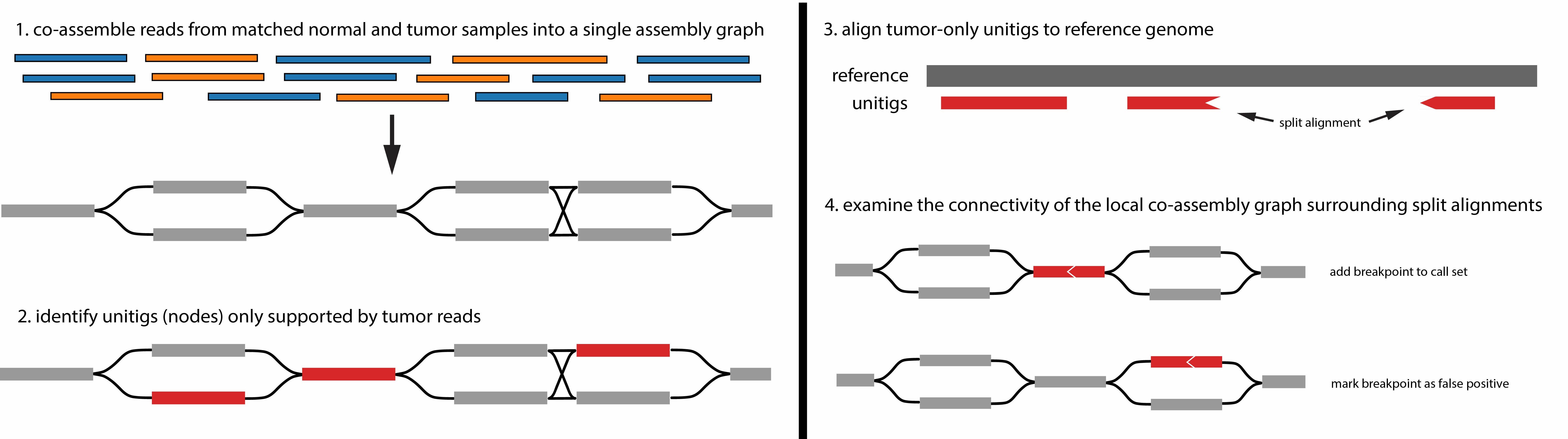
We developed colorSV, a long read-based method for calling large-scale structural variations (SVs). colorSV co-assembles reads from matched tumor-normal samples and examines the local topology of the joint assembly graph to identify true somatic breakpoints. Our method is the first somatic SV calling method that uses a co-assembly approach, as well as the first SV caller that identifies variants by examining characteristics of the assembly graph itself. We demonstrated near-perfect precision and sensitivity for calling translocations on the COLO829 cell line, outperforming four existing somatic SV callers (Severus, Sniffles2, nanomonsv, and SAVANA) in both metrics.
Detecting Natural Selection in Ancient Populations Using Models of Admixture
[preprint] [seminar] [summary thread] [code]
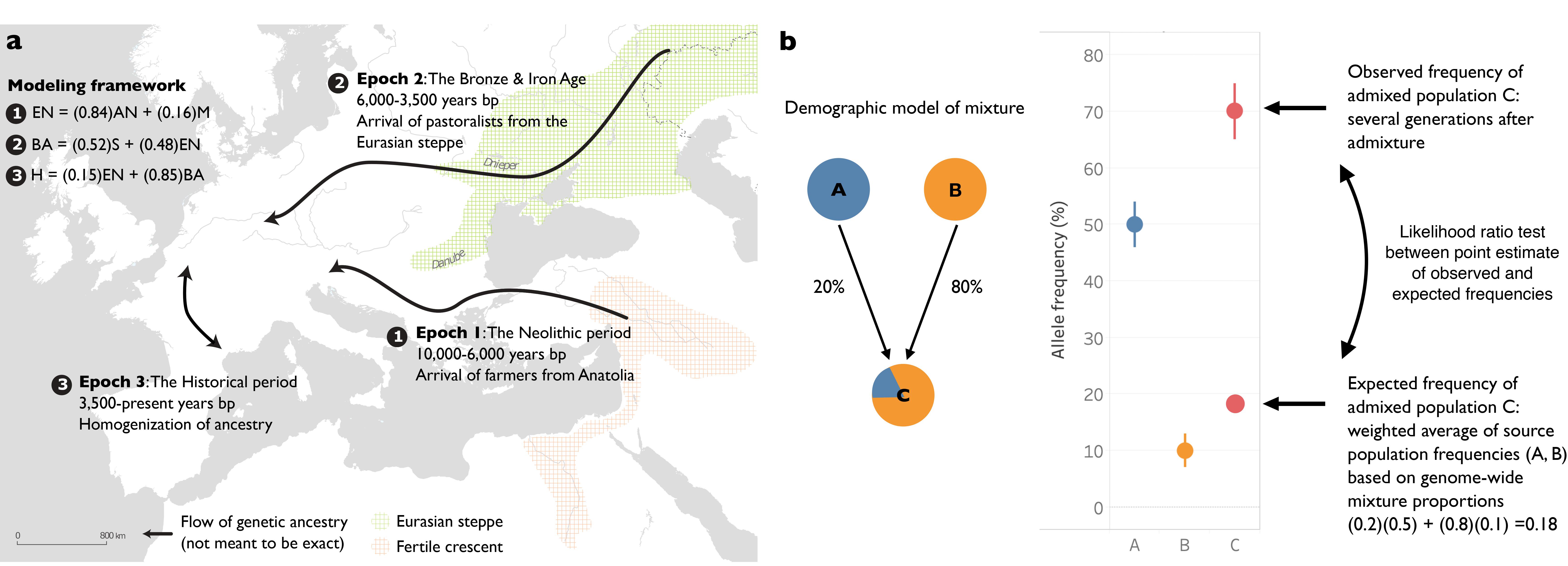
We analyzed 1,291 ancient DNA samples from Europe to identify signals of selection in the context of major environmental changes over the past of 10,000 years. Using models of population admixture, we detected loci where allele frequencies changed beyond expectation. This approach allowed us to stratify our data and date our results to three different epochs: the Neolithic, the Bronze Age, and the Historical periods. We also combined our selection statistic with GWAS summary statistics from Biobank Japan to create a test for polygenic selection on complex traits with increased robustness to population stratification.
Evaluating ECCOv4r4 Currents in the Pacific Equatorial Undercurrent
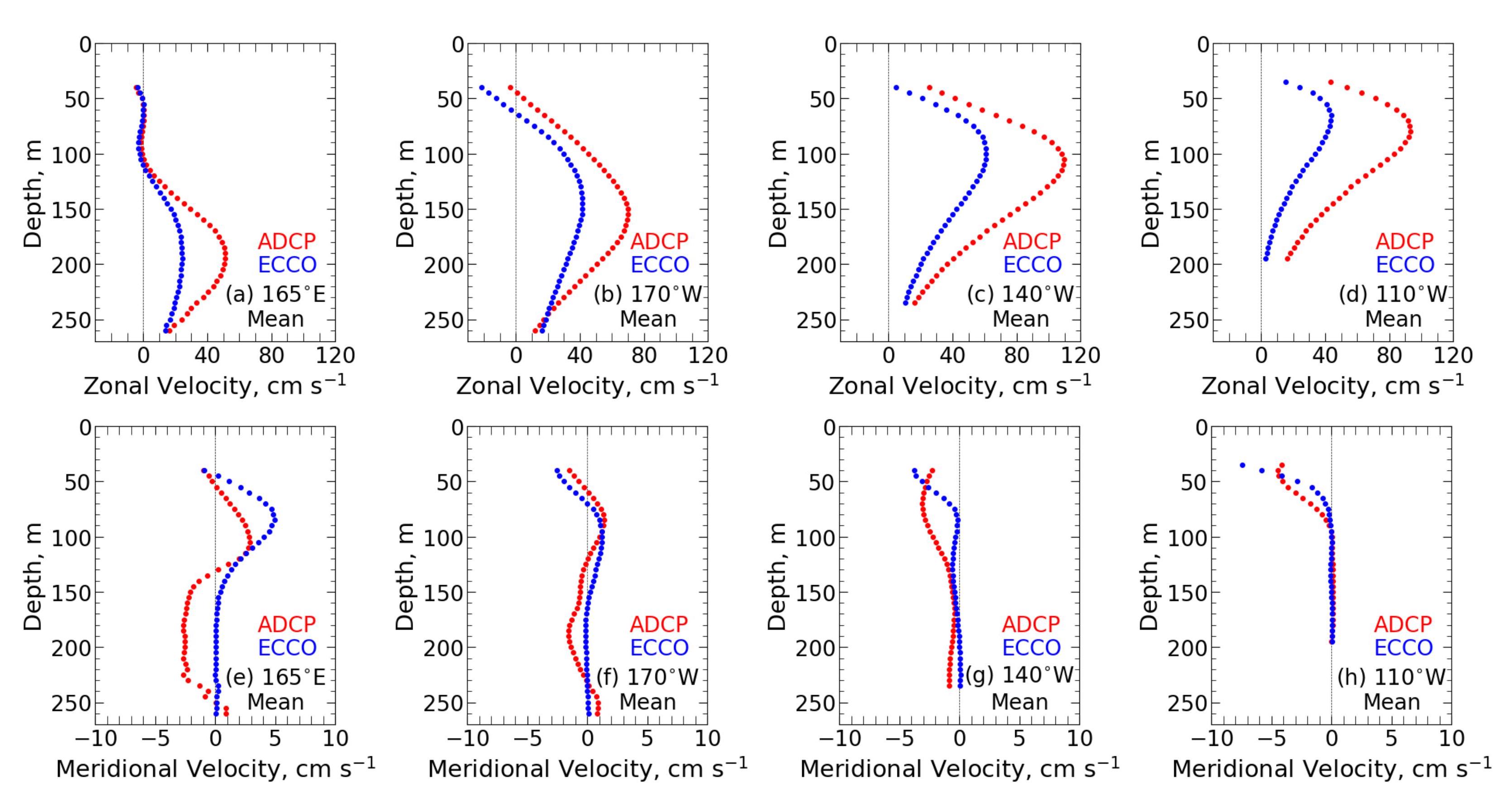
We compared currents in the Pacific Equatorial Undercurrent generated by the ECCO Consortium model to in-situ moored acoustic Doppler current profiler measurements. We compared current characteristics (including means and standard deviations of velocities, core speeds, thicknesses, transports per unit width, extreme values, and Richardson numbers) at four equatorial sites from 1995 through 2010. We also performed spectral analysis, examined El Niño events, and evaluated the impact of assimilated in-situ data.
Feel-the-Force: Chemical Simulation Software with Haptic Feedback
[poster] [code] [lab project page]
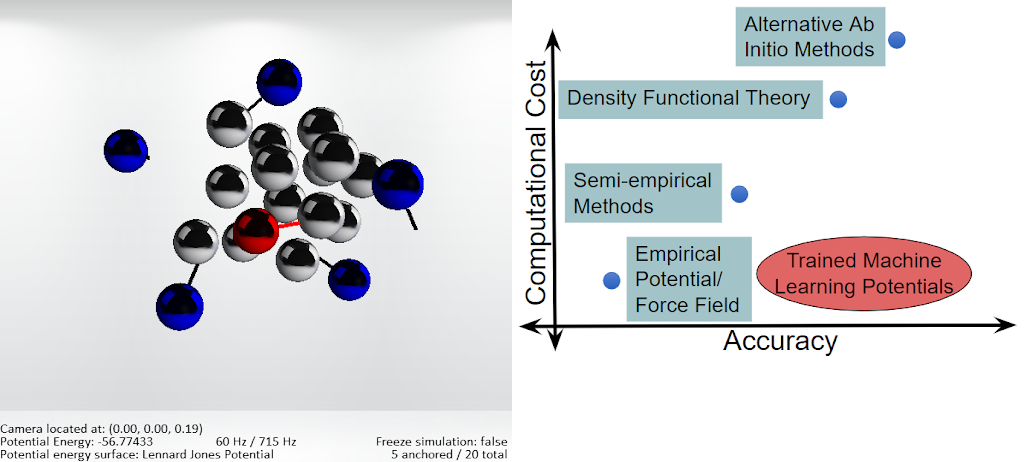
We wrote software to simulate and manipulate chemical systems for a haptic device that provides physical feedback and control of the atoms, allowing users to gain intuition about atomic interactions. The program calculates potential energies and forces using the positions of the atoms, then scales the forces to be translated to a haptic device. My work included editing the Atomistic Machine-learning Package for integration of machine-learning generated potential energy surfaces. I also implemented the Morse potential for potential energy surface calculation, added mouse control of the atoms, and implemented the ability to save and load atomic configurations.
Benchmarking Negative Curvature Solutions to High-Dimensional Newton’s Method
[code]
We implemented different solutions to the negative curvature problem for high-dimensional Newton's Method to determine which method is most efficient for optimizing the Lennard-Jones 38 cluster. These solutions included different methods for selecting a shift parameter used in the calculation of Newton’s Method’s step, using a trust radius, and taking the absolute value of negative eigenvalues when calculating each step. The performance of each method was measured as the average number of force calls needed to optimize the same 100 randomly generated Lennard-Jones 38 configurations.